- Title
-
Zebrafish Naked1 and Naked2 antagonize both canonical and non-canonical Wnt signaling
- Authors
- Van Raay, T.J., Coffey, R.J., and Solnica-Krezel, L.
- Source
- Full text @ Dev. Biol.
|
Alignment and expression of zebrafish nkd1 and nkd2. (A) Alignment of zebrafish Nkd1 and Nkd2a amino acid sequence with human Nkd1 and Nkd2. Red boxes identify identical residues, while yellow boxes identify similarities. Numbering refers to the amino acid sequence of zebrafish Nkd1. Overlying bars define the different conserved domains. Myr, myristoylation sequence; NHR1, naked homology region 1; EF hand, identifies the homologous region in D. melanogaster Nkd that is predicted to bind zinc. The zinc-binding residues are denoted with an asterisks according to Zeng et al. (2000); NHR2, naked homology region 2; TTB, TGFα tail binding domain that is found in human Nkd2 (Li et al., 2004); His, Histidine rich domain. (B–L) Whole-mount in situ hybridization analysis of nkd1 expression. (B) nkd1 expression is not detected in 16 cell stage, Pre-MBT embryos. (C) Expression of nkd1 is detected in the presumptive organizer region (arrowhead) at dome stage. (D) At 50% epiboly, nkd1 expression is in the entire blastoderm margin and by shield stage is reduced in the shield (E), only to be turned on in the axial mesendoderm (F). The solid line through the shield marks the plane of section shown in panel F. In panels F and H, the dashed line demarcates the ectodermal epiblast (e) and the mesendodermal hypoblast (h) layers and the arrow depicts route of cell movements during internalization. (G) At 75–80% epiboly, expression is observed in the margin and in the axial mesendoderm (arrowhead). The arrow identifies the plane of section in panel H. (I) Tailbud (TB) stage. Mid-hindbrain boundary (mhb) is identified with an arrowhead. (J) In early somitogenesis, arrowhead identifies the mhb and the arrow is pointing to segmented expression in the hindbrain. (K) 10 somite stage, expression is observed in the mhb (arrowhead), hindbrain (arrows), developing somites (*) and in the presomitic mesoderm (bracket). (L) At 1 dpf, expression continues in the mhb (arrow) but is now also observed in the developing eye (arrowhead). (M–R) Whole-mount in situ analysis of nkd2 expression. (M–Q) nkd2 expression is ubiquitous. At 1 dpf, expression is confined to the anterior CNS and the posterior tail. (M) 16 cell stage; (N) dome stage; (O) 50% epiboly; (P) 75–80% epiboly; (Q) early somitogenesis stage; (R) 1 dpf. (S) Relative RNA levels determined by qRT-PCR for nkd1. Levels are normalized to 1 dpf (16 cell = 0.15±0.43; Dome = 0.48±0.21; Shield = 1.92 ± 1.37; 1 dpf = 1±0.56). Error bars represent standard deviation. (B, M, N) lateral view, animal pole is to the top; (C, G, N, P) dorsal view, animal pole to the top; (D, O) lateral view, dorsal is to the right, animal pole to the top; (E) animal pole view, dorsal is to the right; (I, J, L, Q, R) lateral view, anterior is to the left; (K) flat mount view, anterior is the to the left. (F, H) Sagittal sections through regions identified in panel E (line) and panel G (arrow). EXPRESSION / LABELING:
|
|
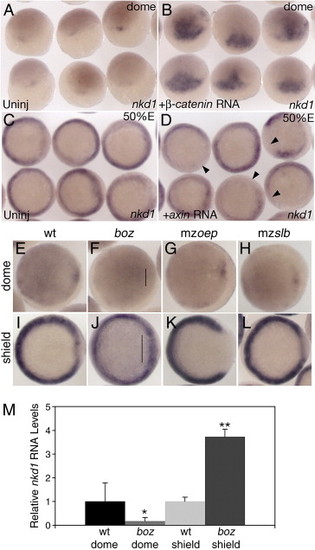
Nkd1 is a target of canonical Wnt/β-catenin signaling. Overexpression of constitutively active β-catenin RNA results in ectopic expression of nkd1 at dome stage (B) compared to uninjected controls (A). Overexpression of axin RNA reduces endogenous nkd1 expression in the mediolateral margin at 50% epiboly (D) compared to uninjected controls (C). Arrowheads point to domains of reduced or absent nkd1 expression. (E–L) Expression of nkd1 at dome stage (E–H) and shield stage (I–L) in wild-type (E, I) and different mutant backgrounds: (F, J) bozm168, (G, K) maternal zygotic one-eyed pinhead (mzoeptz57) and (H, L) maternal zygotic silberblick (mzslbtz215). Note lack of nkd1 expression at dome stage in boz mutants (line in panel F), but an increase in expression in the dorsal region at shield stage (line in panel J). (A, B) dorsal view, animal pole is to the top. (C, D) Animal view. (E–L) Animal view, dorsal is to the right. While there is no change in nkd1 expression at dome stage in mzoep (G), there is a dramatic reduction in nkd1 expression in the dorsal region at shield stage (K). There is no observable difference in nkd1 expression in mzslb compared to wild type (H, L). (M) Relative qRT-PCR values for nkd1 in wild-type and boz embryos at dome and shield stage. Values are normalized to wild-type for both dome and shield stage (wt dome = 1±0.78; boz dome = 0.17±0.15; wt shield = 1±0.18; boz shield = 3.73±0.32). Error bars represent standard deviation (Students t-test, *, p < 0.01; **, p < 0.001). |
|
Nkds inhibit early canonical Wnt/β-catenin signaling. (B) nkd1 or nkd2a injected within 7 min of fertilization and assayed at sphere stage for mkp3 expression. (A) Uninjected siblings. (D) Injection of either nkd1 or nkd2a within 7 min of fertilization and assayed at 50% epiboly for gsc expression. Arrowheads identify embryos with expanded gsc expression and an asterisk identifies embryos with a decrease in gsc expression. (C) Uninjected siblings. (E) Injection of nkd2 MO at 1–2 cell stage and assayed at sphere stage for mkp3 expression. (F) nkd2 splice site MO control. This data is quantified in panel L. (H) nkd1 MO injected at the 1–2 cell stage and assayed at 30% epiboly for gsc expression. (G) Uninjected siblings. (I) nkd2 MO injected embryos at 1–2 cell stage and assayed for gsc expression at 30% epiboly. (J) A combination of nkd1 MO and nkd2 MOs were injected at the 1–2 cell stage and assayed at 30% epiboly for gsc expression. (K) A graph of the arc of gsc expression at 30% epiboly under different conditions. Error bars represent standard deviation (Students t-test, *, p < 0.05; **, p < 0.001; ***, p < 0.0001). n = number of injected embryos. (A–F) Dorsal view; (G–J) Animal view, dorsal is down. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Nkds inhibit late canonical Wnt/β-catenin signaling. (A, B) Standard 1 to 2 cell overexpression of nkd1 or nkd2a RNA significantly expands gsc expression at 50% epiboly (B) compared with uninjected control siblings (A). (C) Injection of nkd1 MO, nkd2 MO and nkd1 or nkd2a RNA that is insensitive to the MOs results in an even larger expansion of gsc expression at 50%. (A–C) Dorsal view. This data is quantified in panel D. Error bars represent standard deviation (Students t-test, *, p < 0.05; ***, p < 0.0001). EXPRESSION / LABELING:
|
|
Nkds can antagonize ectopic Wnt8. (A) Overexpression of Wnt8 in wild-type (wt) embryos results in an eyeless phenotype. (B) Co-expression of wnt8 and nkd1 or nkd2 results in a near complete suppression of the Wnt8 effect. This is quantified in panel C. (D–G) boz mutants have ectopic Wnt8 activity in the dorsal mesoderm, which results in a reduction of axial tissue as demonstrated by reduced ntl expression (D), or in the physical absence of a notochord (F). The presumptive neuroectoderm is labeled with six3b which is also reduced in boz mutants (D). Injection of nkd1 or nkd2a into boz embryos suppresses the deficiency of ntl expression in the notochord (E) and of a notochord at 24 hpf (G). Error bars represent standard deviation (Students t-test; ***, p < 0.0001). (A, B, F, G) Lateral view, anterior to the left; (D, E) dorsal view. |
|
Nkds inhibit non-canonical Wnt/PCP signaling. (A) Wild-type embryos have a cyclopia index (CI) of 1, while slb mutants have a CI of 2.80 ± 0.46 (B). (C) slb embryos injected with nkd1 or nkd2a RNA have an increase in cyclopia with a CI of 3.77 and 3.08 respectively. See text for details. (D–I) Analysis of prechordal plate position. (D) Wild-type embryos have a narrow neural plate (double headed arrow) and a prechordal plate that has fully extended past the neuroectoderm and non-neural ectoderm boundary (black arrowhead). (E) slb mutant embryos exhibit a wider neural plate due to impaired C&E (double headed arrow) and reduced extension of the axial mesendoderm (black arrowhead). (F) Injection of wnt11 RNA into slb embryos can rescue the C&E defects in slb embryos. (G) Overexpression of nkd1 or nkd2a RNA in slb embryos exacerbates the axial mesendoderm extension defect. (H) Co-expression of wnt11 with nkd1 or nkd2a results in a phenotype that resembles the nkd overexpression phenotype arguing that Nkds can block Wnt11 signaling. This data is semi-quantified in (I) where moderate is defined as ≤ ∼50% of the hgg expression domain is within the dlx3 expression domain and severe is defined as > ∼50% of the hgg expression domain is within the dlx3 domain. (A–C) 2 dpf, (A) dorsal view, (B, C) ventral views. (D–H) 2–3 somites, anterior view, dorsal is to the top, prechordal plate marked by hgg (red), neuroectoderm-non neural ectoderm boundary marked by dlx3 (dark purple), developing somites (out of view) marked by dlC for staging confirmation. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Reduction of Nkd1 can suppress the axial C&E defect in slb. (A) Uninjected slb mutant. (B) slb embryo injected with nkd1 MO results in a suppression of the axial C&E defect observed in uninjected slb embryos, which is quantified in (J). Note that the convergence of the neural plate remains unchanged (K). In contrast, injection of nkd2 MO exacerbates both the axial C&E (C, J) and the neural plate convergence phenotype in slb mutants (C, K). (D–F) Dorsal view, animal pole to the top of gsc expression at 50% epiboly in slb mutants which are uninjected (D), injected with axin RNA (E) or axin RNA and nkd1 MO (F). The arc of gsc expression is quantified in panel L. (G–I) slb embryos from the same clutch as (D–F) analyzed for percent of axial extension. Anterior view, dorsal to the top of uninjected (G), axin RNA injected (H) and axin RNA and nkd1 MO injected embryos (I). The percentage of axial extension is quantified in panel M and the width of the neural plate is quantified in panel N. (A–C, G–I) 2–3 somite stage. The percentage of complete axial extension is calculated by measuring the proportion of total hgg expression that is over and past the dlx3 expression domain. One hundred percent would be defined as complete extension, as observed, for example, in wild-type (Fig. 7D). Markers are as described in Fig. 7. Error bars represent standard deviation (Students t-test, *, p < 0.05; **, p < 0.01; ***, p < 0.0001). EXPRESSION / LABELING:
PHENOTYPE:
|
Reprinted from Developmental Biology, 309(2), Van Raay, T.J., Coffey, R.J., and Solnica-Krezel, L., Zebrafish Naked1 and Naked2 antagonize both canonical and non-canonical Wnt signaling, 151-168, Copyright (2007) with permission from Elsevier. Full text @ Dev. Biol.