- Title
-
Mutant analyses reveal different functions of fgfr1 in medaka and zebrafish despite conserved ligand-receptor relationships
- Authors
- Yokoi, H., Shimada, A., Carl, M., Takashima, S., Kobayashi, D., Narita, T., Jindo, T., Kimura, T., Kitagawa, T., Kage, T., Sawada, A., Naruse, K., Asakawa, S., Shimizu, N., Mitani, H., Shima, A., Tsutsumi, M., Hori, H., Wittbrodt, J., Saga, Y., Ishikawa, Y., Araki, K., and Takeda, H.
- Source
- Full text @ Dev. Biol.
|
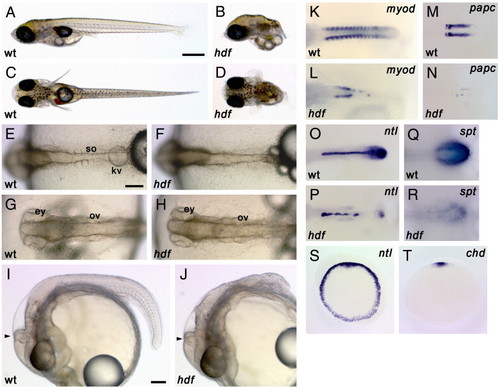
Morphology and gene expression in headfish mutant embryos. (A–J) External morphology. Wild-type (A, C) and hdf mutant (B, D) embryos at hatching stage (st. 39). Lateral view (A, B) and dorsal view (C, D). Mutant embryos lack most of the posterior part of the body. On the other hand, head development appears normal. Wild-type (E, G) and hdf mutant (F, H) embryos at 4-somite stage (st. 20). Dorsal view of the tail (E, F) and head (G, H). Mutant embryos have a midline structure but fail to form somites. Lateral view of wild-type (I) and hdf mutant (J) embryos at 30-somite stage (st. 28). hdf mutant embryos show agenesis of the posterior structures but have a morphologically normal head, including the eyes, ventricles and MHB (arrowhead). ey, eye; kv, Kuppfer's vesicle; ov, otic vesicle; so, somite. Scale bars: (A) 500 μm; (E) 100 μm; (I) 100 μm. (K–T) Gene expression patterns. Wild-type (K, M, O, Q) and mutant (L, N, P, R) embryos during early segmentation stages. (K, L) The expression of myod, a somite differentiation marker, is observed in each somite in a wild-type embryo, in a bilateral regular stripe pattern (K), whereas it is severely reduced in the mutant embryos (L). (M, N) The expression of papc, a presomitic mesoderm marker, is lost and only faint expression is observed in the mutant (N). (O, P) The expression of ntl, a pan-mesodermal marker. ntl expression is detected in the notochord and tailbud in wild-type (O), but is down-regulated in tailbud and exhibits a interrupted pattern in midline of the mutant embryos (P). (Q, R) The expression of spt, a marker for the tailbud. While spt is expressed in the tailbud of the wild-type embryo (Q), it is considerably weaker in the mutant embryo (R). (S, T) Early gastrula stage. Animal-pole views are shown. ntl is expressed uniformly in the blastoderm margin of all sibling embryos obtained from a heterozygous mating (S) and chordin, a dorsal mesodermal marker is detected in the dorsal region of all sibling embryos (T). Each genotype is indicated as wt, wild-type and hdf, headfish. |
|
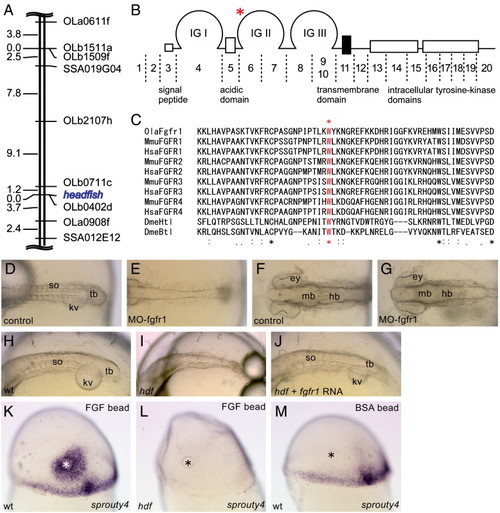
Genetic mapping and identification of the headfish mutation. (A) The hdf locus is roughly mapped in the middle of medaka LG9, near the EST marker OLb0402d. (B) Structure of FGFR1 and its corresponding exons. Medaka fgfr1 consists of at least 20 exons. A G-to-C point mutation was found in exon 6, indicated by a red asterisk. The mutation causes an amino acid substitution W181C in the second immunoglobulin (IG) domain. (C) The mutated tryptophan residue is conserved among all of the FGFRs from Drosophila to human, indicating its essential role for the FGFR function. (D–G) Morpholino knock-down of fgfr1. Dorsal views of the trunk (D, E) and head (F, G) regions of control (D, F) and morphant (E, G) embryos are shown. Note that MO injection phenocopies hdf mutants. Anterior to the left. (H–J) Rescue of the mutant phenotype by injection of RNA encoding wild-type Fgfr1. Oblique dorsal views of the trunk region of control (H), hdf mutant (I) and RNA-injected hdf mutant (J) are shown. Note that the injected hdf mutant forms somites (J). Anterior to the left. (K–M) Responsiveness to exogenous FGFs. Mid-gastrula embryos transplanted with FGF8-beads on the dorsal side were stained with sprouty4 probe. Lateral views of wild-type (K), hdf mutant (L) and control wild-type (M) embryos are shown. Animal pole to the top. The control embryo was transplanted with a BSA-soaked bead (M). In addition to endogenous expression in the marginal and dorsal mesoderm, sprouty4 expression is induced around the FGF8-beads (K), whereas the FGF8-beads fail to induce sprouty4 in the hdf mutant (L). |
|
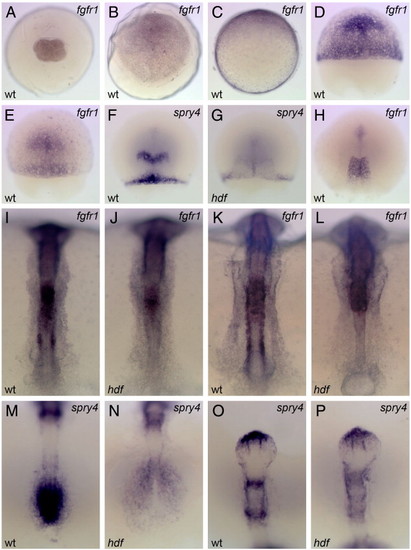
Embryonic expression of medaka fgfr1 and its down-stream target, sprouty4. Probes used are on the top right corner. wt, wild-type and hdf, fgfr1hdf mutant embryos. Animal-pole views (A–B) and dorsal views (E–P; animal-pole or anterior to the top) are shown. Maternal expression of medaka fgfr1 is detected ubiquitously at st. 4 (4-cell) (A). At st. 13 (early gastrula), fgfr1 is expressed ubiquitously with higher levels on the dorsal side (to the top; C). During gastrulation, fgfr1 is expressed in the presumptive head region with the continued expression in the margin (D, st. 15; E, st. 15+). The expression of sprouty4 is observed in the presumptive MHB and dorsal margin in a part of the sibling embryos (F), but in some siblings this expression is undetectable (G). At the end of gastrulation, fgfr1 is expressed in the trunk region (H). At the bud stage (I) and the early segmentation stage (K), fgfr1 is expressed in the trunk, paraxial mesoderm, somite and presomitic mesoderm. In the neural tube, weak ubiquitous expression with high levels in the hindbrain is observed in the wild-type embryo (I, K). In the mutant, fgfr1 expression is lost in the paraxial mesoderm, somites and presomitic mesoderm, due to a lack of these tissues, whereas the expression in the anterior portion remains unchanged (J, L). At these stages, sprouty4 is expressed in the tailbud (M), the anterior tip of telencephalon, MHB and r4 (O). The sprouty4 expression is lost in the tailbud and reduced in the mutant (N, P), except for the telencephalon. |
|
Activity of fgfr1 is transiently required for rhombomere patterning. Dorsal view and lateral view of wild-type (A–C, G–J) and fgfr1hdf (D–F, K–N) embryos. At st. 18, krox20 expression in the r5 is lost in the mutant, whereas the expression in r3 is not affected (A, D). The expression in r5 is restoring at st. 20 in the mutant (B, E), and is recovered at st. 22 (C, F). The expression of pax6 in forebrain and hindbrain is normal in the mutant (G, H, K, L). The expression of pax2 in MHB (I, M) and shh in prechordal plate and floor plate (J, N) are not affected in the mutant. |
|
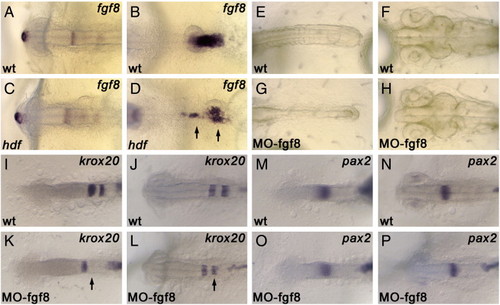
The expression of fgf8 and the phenotype of fgf8-morphant. Medaka fgf8 is expressed in the anterior tip of the forebrain, MHB, hindbrain and tailbud (A, B). The expression of fgf8 is largely unaffected in the fgfr1hdf mutant, except for the expression in tailbud (C, D; arrows). Injection of fgf8 morpholino causes somite-less phenotype, phenocopying the fgfr1hdf mutant phenotype (E, G). Same as fgfr1hdf, morphology of the brain including MHB is normal in the fgf8 morphant (H). The expression of krox20 and ntl in wild-type (I, J) and fgf8 morphant (K, L). In the morphant, the expression of krox20 in r5 is lost at st. 18, and is recovered at st. 20 (K, L; arrows). The expression of pax2 and ntl in wild-type (M, N) and fgf8 morphant (O, P). The expression of pax2 in MHB is not affected in the morphant. |
Reprinted from Developmental Biology, 304(1), Yokoi, H., Shimada, A., Carl, M., Takashima, S., Kobayashi, D., Narita, T., Jindo, T., Kimura, T., Kitagawa, T., Kage, T., Sawada, A., Naruse, K., Asakawa, S., Shimizu, N., Mitani, H., Shima, A., Tsutsumi, M., Hori, H., Wittbrodt, J., Saga, Y., Ishikawa, Y., Araki, K., and Takeda, H., Mutant analyses reveal different functions of fgfr1 in medaka and zebrafish despite conserved ligand-receptor relationships, 326-337, Copyright (2007) with permission from Elsevier. Full text @ Dev. Biol.