- Title
-
Baf60c is a nuclear Notch signaling component required for the establishment of left-right asymmetry
- Authors
- Takeuchi, J.K., Lickert, H., Bisgrove, B.W., Sun, X., Yamamoto, M., Chawengsaksophak, K., Hamada, H., Yost, H.J., Rossant, J., and Bruneau, B.G.
- Source
- Full text @ Proc. Natl. Acad. Sci. USA
|
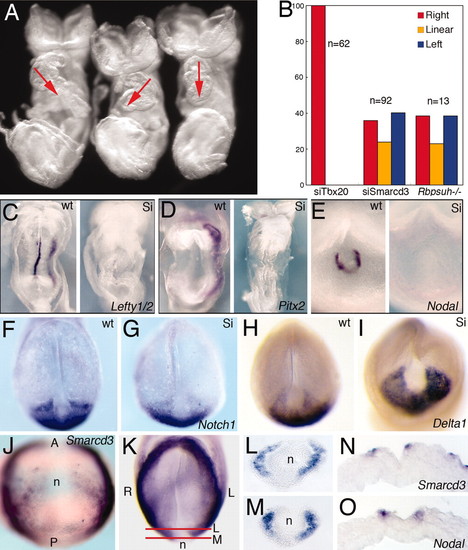
Baf60c regulates LR asymmetry at the node. (A) Smarcd3 knockdown caused randomized heart looping. Red arrows indicated the direction of heart looping. (B) Histogram shows the percentage of control (siTbx20), Smarcd3 knockdown (siSmarcd3), and Rbpsuh -/- mutant embryos with rightward, linear, and leftward heart morphologies. (C–E) Expression of LR regulators in wild-type (wt) and Smarcd3 knockdown (Si) embryos. Shown are Lefty1/2 (C), Pitx2 (D) at E8.25, and Nodal (E) at E7.75. (F–I) Decreased expression of Notch1 and increased expression of Dll1 in siSmarcd3 embryos. (J–O) Smarcd3 expression in perinodal cells and coexpression with Nodal by in situ hybridization on consecutive sections at E7.75 (O). Red lines in K show the plane of sections shown in L and M. |
|
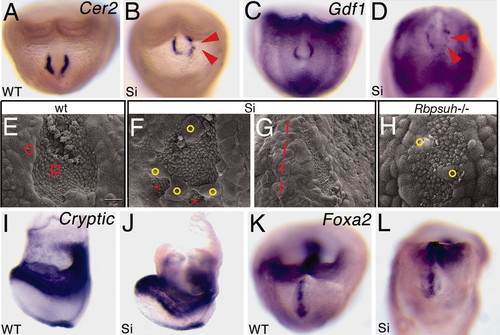
Abnormal node morphology in Smarcd3 knockdown embryos. (A–D) Expression of Cer2 and Gdf1 in wild-type and Smarcd3 knockdown (Si) embryos. Red arrowheads show decreased or missing expression. (E–K) Scanning electron microscopy shows high magnification of the node in wild type (E), abnormal node morphology in Smarcd3 knockdown (F and G), and Rbpsuh -/- (H) embryos at E7.75. c, crown cells; p, pit cells. Smarcd3 knockdown results in abnormal node morphology, with separated pit cells (F, red asterisks) and abnormally migrated crown cells (yellow circles). Some Smarcd3 mutants also have abnormal leftward shifted nodes in addition (G, red line shows embryonic midline). Abnormal crown cells were seen in Rbpsuh -/- embryos (H, yellow circles). (I–L) Expression of Cryptic and Foxa2 in wild-type and Smarcd3 knockdown (Si) embryos. |
|
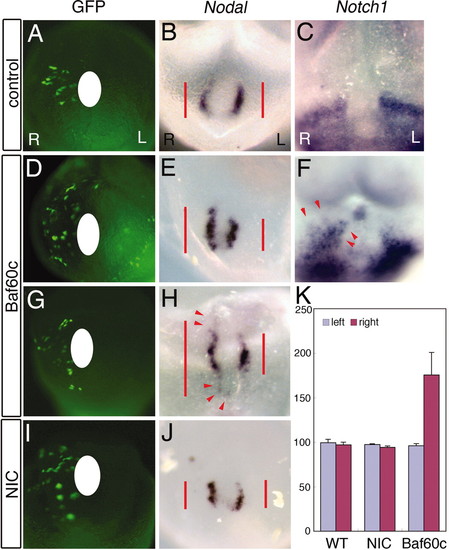
Baf60c is sufficient to induce Nodal. Cultured embryos transfected with an EGFP expression construct and Baf60c, activated Notch (NIC), or Nodal expression constructs, or EGFP vector alone (control) were cultured for 15 h. (A, D, G, and I) EGFP fluorescence, showing transfected cells. (A–C) Control studies showed normal/bilateral nodal (B) at the node and Notch1 (C) expression around the node. (D–H) Smarcd3-transfected embryos: ectopic Nodal induction along the midline was seen (E and H), and expansion of Notch1 expression was also observed (F, red arrowhead). (I and J) Overexpression of NIC enhanced Nodal expression. (K) Histogram shows the length (in millimeters) of Nodal expression on the node between the left side (light purple, control) and the right side (light blue, injected experiments, n = 5). |
|
Smarcd3 is required for LR axis specification in zebrafish. (A–E) Expression of smarcd3 (A–C and E are lateral views, anterior to left, and D is a caudal view, dorsal to top). (A) During gastrulation. (B) Expression in the developing tailbud. (C and D) Early somitogenesis. (E) Mid-somitogenesis. Knockdown of smarcd3 randomizes direction of heart looping in embryos 40 h after fertilization (F–H). A, atrium; V, ventricle. In H, red bars show normal looping and blue bars show reversed looping. Heart looping is partially rescued by smarcd3 RNA. (I–L) Altered expression of lefty1 (lft1) in the diencephalon and lefty2 (lft2) in the heart field of 22- to 24-somite-stage embryos. (I) Normal left-sided expression. (J) Right-sided expression. (K) Bilateral expression. (L) Absence of expression. Expression of lft1 in the midline is unaltered. (M–R) Quantification of alterations in asymmetric gene expression patterns in smarcd3 morphants. (M and P) southpaw (spaw) expression in the LPM (19- to 21-somite stage). (N and Q) lft1 expression. (O and R) lft2 expression. L, left-sided; R, right-sided; B, bilateral; A, absent. EXPRESSION / LABELING:
PHENOTYPE:
|

Unillustrated author statements EXPRESSION / LABELING:
PHENOTYPE:
|