- Title
-
Differential regulation of the zebrafish orthopedia1 gene during fate determination of diencephalic neurons
- Authors
- Del Giacco, L., Sordino, P., Pistocchi, A., Andreakis, N., Tarallo, R., Di Benedetto, B., and Cotelli, F.
- Source
- Full text @ BMC Dev. Biol.
|
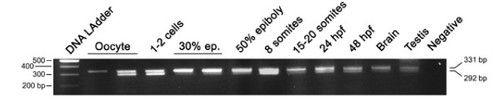
Developmental RT-PCR analysis of the otp1 transcripts. Ethidium Bromide-stained agarose gel of RT-PCRs performed using the zotpS and zotpRv primers: DNA Ladder (lane 1), oocyte (lanes 2 and 3), 1-2 cell stage (lane 4), 30% epiboly (lanes 5 and 6), 50% epiboly (lane 7), 8 somite stage (lane 8), 15-20 somite stage (lane 9), 24 hpf (lane 10), 48 hpf (lane 11), brain (lane 12), testis (lane 13), and negative control (lane 14) in absence of cDNA. 80% epiboly, 1-2 somites, and 10 somites samples (not shown) resulted positive after the analysis. On the left of the panel is shown the size of the DNA Ladder bands, on the right is indicated the size of the 2 PCR products, 292 bp and 331 bp, corresponding to the short and the long form of the otp1 mRNA, respectively. |
|
Zebrafish otp1 expression in the hindbrain. Anterior is left in all images except for C. All views are dorsal except for E, G and H that are lateral. A, At 11 hpf stage, a narrow transverse stripe encompasses the neural tube in its anterior half. During somitogenesis, a second narrow stripe appears more anteriorly (data not shown). B, Double labelling with otp1 (blue) and krox20 (red) shows that otp1 expression in the hindbrain labels rhombomere 3 (r3) and 5 (r5) at 14 hpf stage. This developmental window coincides with the restriction of the two otp1 stripes toward the lateral margins of the neural tube, and with the early extension of the signal throughout the rhombencephalon. C, Transverse section of a 24 hpf stage zebrafish embryo through the hindbrain shows that these lateral clusters belong to the mantle layer of the ventral alar plate and are placed medially along the dorsoventral axis. D, Following (19 hpf), otp1 signals extend anteriorly, and then posteriorly, in the shape of repeated patterns. E, 24 hpf embryo; otp1 mRNA signal localizes in restricted groups of cells, in a pattern reminiscent of rhombomere segmentation. F, G, otp1 WISH combined with acetylated α-tubulin (TU) immunochemical staining confirms that otp1 transcripts occur in paired clusters within each rhombomere (30 hpf) (open arrowheads in F indicate the axons in the central part of every rhombomere). H, At 34 hpf, otp1 domains have elongated dorsally in parallel columns (vertical dashes in E, F and H, and dashed line in G, indicate rhombomere boundaries and profile, respectively). ce, cerebellum; dap, dorsal alar plate; is, isthmus; n, notochord; ov, otic vesicle; ra, reticulospinal axons; vap, ventral alar plate. EXPRESSION / LABELING:
|
|
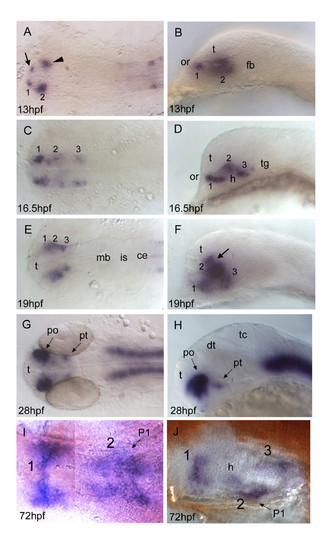
otp1 expression in the forebrain. Anterior is always to the left. Except I, a ventral view, right panels are lateral views and left ones are dorsal. Eyes have been removed in I and J. A, B, About 1 h after early otp1 expression in the hindbrain (11 hpf, see Fig. 4A), a small lateral domain appears at the anterior edge of the central nervous system (arrow) (see also Fig. 6A). Shortly after, a novel center of transcription emerges posterolaterally in the alar plate (arrowhead). C, D, Then, otp1 expression diffuses posteriorly in a segmented fashion with three clusters (1–3) (16.5 hpf). During this developmental phase, spatial rearrangements of the anterior forebrain start occurring as the result of the ventral bending of the neuraxis [50]. Even though intrinsic topological changes in otp1 transcription may also happen, their extents are eventually obscured by morphogenetic dislocations in the neural tube. E, F, At 19 hpf, otp1 positive cells in the middle of longitudinal domains displace dorsally with respect to cells at different axial positions. Consequently, otp1 labelling in the anterior forebrain coalesces in the proximity of the optic recess (arrow). G, H, 28 hpf WISH with a large compacted anterior diencephalic cluster in PO and a small one caudally in PT. I, J, Later in development (72 hpf), otp1 expression is detected in three distinct domains (1–3) in the ventral diencephalon. Unlike early in development, these three clusters are fused along the midline. ce, cerebellum; dt, dorsal thalamus; fb, forebrain; h, hypothalamus; is, isthmus; mb, midbrain; P1, pharyngeal arch 1; po, preoptic area; pt, posterior tuberculum; t, telencephalon; tc, tectum; tg, tegmentum. EXPRESSION / LABELING:
|
|
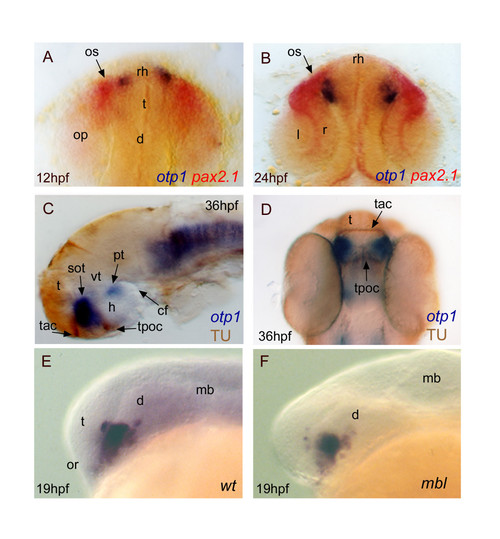
Topology of otp1 transcripts in forebrain compartments. Anterior is top in A, B and D dorsal views, otherwise left and lateral view panels. Eyes have been removed in C. A (12 hpf), B (24 hpf), early otp1 expression foci (blue) occur at the anteromedial edge of pax2.1 (red) pattern in the optic stalks, as shown by double WISH. C, D, Co-labelling with anti-acetylated α-tubulin antibody (brown) and otp1 riboprobe (blue) demonstrates that the anterior otp1 cell cluster is delimited by the supraoptic tracts and those of the anterior and posterior commissures (36 hpf). E, F, mbl embryos are not altered in the dorsal extension of the PO otp1 cluster, if compared with control embryos (19 hpf). cf, cephalic flexure; d, diencephalon; h, hypothalamus; l, lens; mb, midbrain; op, optic primordia; or, optic recess; os, optic stalks; r, retina; rh, rostral hypothalamus; sot, supraoptic tract; t, telencephalon; tac, tract of the anterior commissure; tpoc, tract of the postoptic commissure; vt, ventral thalamus. EXPRESSION / LABELING:
|
|
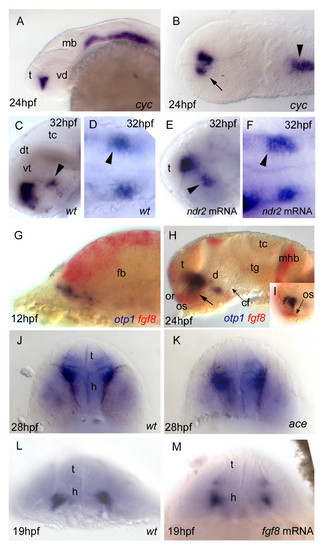
Nodal and FGF signalings are related with otp1 in the diencephalon. Anterior is left in all but J-M panels. J-M panels are frontal views, B, D and F are dorsal, and the remaining ones are lateral. A, B, Nodal signals emanating from mesendoderm and overlaying neuroectoderm represent one of the most important inducing pathways that govern regional patterning of the neural tube. Zebrafish cyclops (cyc) mutant embryos lack a functional floor plate, and indirectly disrupt HH signaling [55, 56, 57, 58]. At 24 hpf, cyc mutants display complete loss of otp1 expression in the PT, and show an asymmetric reduction of PO domains (arrow). Forebrain and hindbrain patterns are closer or fused along the midline (arrowhead). C-F, Accordingly, the essential role of Nodal signaling in otp1 transcription in the posterior hypothalamus is confirmed by microinjection of ndr2 mRNA whereas, compared with controls (C, D), otp1 domain in the PT is expanded (E, F) (arrowhead in C-F indicates the PT-specific otp1 expression) (32 hpf). G-I, Double WISH reveals that fgf8 (red) and otp1 (blue) expression patterns are complementary at the boundary between prospective telencephalon and diencephalon since early somitogenesis stages (G, 5 s).H, At 24 hpf, PO otp1 expression is adjacent to fgf8-positive optic stalks (arrow). I, A high power image of H illustrates the physical proximity of otp1 and fgf8 transcripts. J, K, fgf8 gene activity is abolished in acerebellar (ace) mutant [37]. Examination of an optic section of ace embryos at 28 hpf through the optic area shows that the PO-specific otp1 cluster is expanded radially (K) in comparison with controls (J). L, M, Overexpression of fgf8 induces a reduction of the number of otp1 expressing cells in the PO, whereas this cluster also fails to properly coalesce, likely due to developmental delay (19 hpf). On the contrary, the PT-specific otp1 domain is unaltered (data not shown). cf, cephalic flexure; d, diencephalon; dt, dorsal thalamus; fb, forebrain; h, hypothalamus; mb, midbrain; mhb, midbrain-hindbrain boundary; or, optic recess; os, optic stalks; t, telencephalon; tc, tectum; tg, tegmentum; vd, ventral diencephalon; vt, ventral thalamus. EXPRESSION / LABELING:
|
|
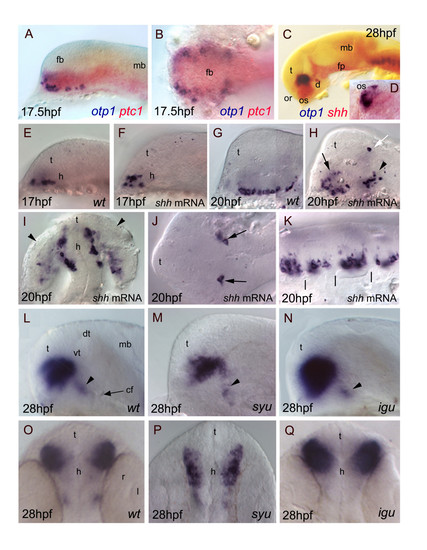
Relationships between otp1 and HH signaling in the diencephalon. Anterior is to the left in all panels except for D, I, O-Q where anterior is up. The latter panels, B and J are dorsal images, while the remaining ones are lateral views.A-D, Relative distribution of otp1 and shh signaling factors, i.e. shh and its receptor patched-1 (ptc1). A, B, Co-staining of ptc1 (red) and otp1 (blue) mRNAs at 17.5 hpf stage reveals that otp1 expression is included in the shh-pattern, where they are placed at the ventrolateral margins. C, D, Likewise, double labelling of shh (red) and otp1 (blue) at 28 hpf shows coexpression near the optic stalks, as seen in the close-up (D). E-K, Overexpression of synthetic shh mRNA at 1–2 cell stage induces dorsalization of otp1 positive cells. E, F, Since early otp1 expression, forced shh expression induces an initial dorsal displacement at the rostral edge of the diencephalon, without significant alteration of cell number (17 hpf). G, H, A comparable number of otp1 positive cells in both the PO (arrow) and the PT (arrowhead) are dorsalized at later stages of brain development (20 hpf). I, shh overexpressing embryos shown in H display abnormal expression in the optic vesicle (arrowheads). J, In addition, ectopic expression of otp1 is consistently found in two symmetrical cells in the dorsal thalamus (open arrow in H, arrows in J). K, The repeated pattern of otp1 expression in the hindbrain is maintained, but it now appears as a single median cluster per each rhombomere, and it does not extends dorsally (vertical dashes correspond to rhombomere boundaries; compare with Fig. 4E, F-H). L-Q, otp1 expression in 28 hpf zebrafish mutants lacking shh (syu) and the HH-inhibitor Dzip-1 (igu). Labelling of PO cells indicates that the size of the otp1 cluster is (M, P) reduced in syu and (N, Q) enlarged in igu as well, if compared with (L, O) control embryos. In both mutants, no significant alteration of the otp1 cluster in PT is observed (arrowheads in L-N). cf, cephalic flexure; d, diencephalon; dt, dorsal thalamus; fb, forebrain; fp, floor plate; mb, midbrain; l, lens; or, optic recess; os, optic stalks; r, retina; t, telencephalon; vt, ventral thalamus. EXPRESSION / LABELING:
|
|
Synergicistic effects of HH and FGF signaling on otp1 expression in the PO cluster. Anterior is left and dorsal is up in all panels. Eyes have been removed in A. All embryos are 28 hpf and have been double stained with otp1 riboprobe and anti-acetylated α-tubulin (TU) antibody. Compared with the wild-type PO (A), the anterior otp1 cluster is expanded and reduced, respectively, in ace (B) and syu (C), but its normal size is rescued in double ace; syu mutant embryos (D). cf, cephalic flexure; dt, dorsal thalamus; h, hypothalamus; po, preoptic area; pt, posterior tuberculum; sot, supraoptic tract; t, telencephalon; tac, tract of the anterior commissure; tpoc, tract of the postoptic commissure; vt, ventral thalamus. EXPRESSION / LABELING:
|
|
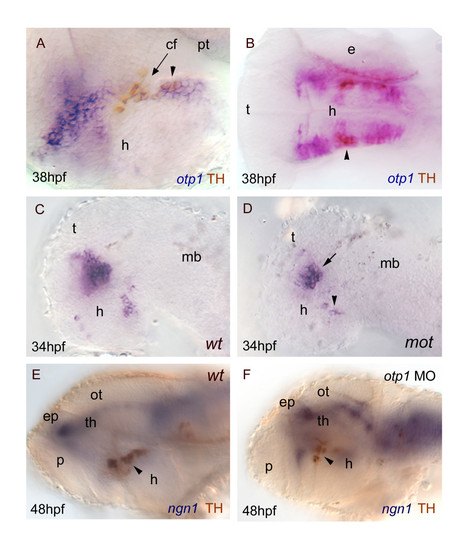
otp1 functions and CA neuron differentiation. Anterior is left and dorsal is up (except for B, dorsal view) in all panels. Eyes have been removed. A, B, anti-TH antibody labels the PT diencephalic CA neurons at 38 hpf [65]. Co-labelling with otp1 is evident in a fraction of TH-positive neuroblasts in the PT (arrowhead). C, D, motionless (mot) embryos display a severe reduction of cell numbers across all DA populations of the zebrafish brainstem, including the diencephalic ones (34 hpf) [63]. Herein, absence of mot functions drastically inhibits the differentiation of otp1 positive cells, more in the PT (arrowhead) then in the PO (arrow). E, F, microinjection of otp1 morpholino oligonucleotide lowers the number of TH-labelled DA neuroblasts in the PT (arrowhead) of 48 hpf embryos that were labelled with neurogenin1 (ngn1), a marker of specific neuronal populations in the forebrain. cf, cephalic flexure; e, eye; ep, epiphysis; h, hypothalamus; mb, midbrain; ot, optic tectum; p, pallium; th, thalamus. |