- Title
-
Zebrafish cypher is important for somite formation and heart development
- Authors
- van der Meer, D.L., Marques, I.J., Leito, J.T., Besser, J., Bakkers, J., Schoonheere, E., and Bagowski, C.P.
- Source
- Full text @ Dev. Biol.
|
(A) Genomic organization of cypher and the 13 identified splice variants from zebrafish heart and skeletal muscle. Shown on top is a schematic for the genomic structure of the zebrafish cypher gene which is found on chromosome 13 and spans a region of 80.125 bp. It consists of 18 exons with exon 4 and 6 containing the ZM motifs. Shown below are thirteen different splice variants (all sequences have been submitted to Genbank). The cypher1 splice variants (containing exon 4) were only found in heart tissue, for the other variants, it is indicated whether they have been found in skeletal muscle (labeled “muscle”) or heart or in both tissues. The cypher1ζ form shows similarity to the mouse Cypher2c splice form and cypher2γ shows similarity to the murine Cypher2s. All other splice forms are novel, and similar splice forms from other species have not been described yet. (B) Alignment of zebrafish cypher2γ to human ZASP and mouse Cypher2s and a species comparison of the ZM motif containing exons 4 and 6. Shown on top is the comparison of the amino acid (aa) sequences of the three homologous splice variants from zebrafish (cypher2γ submitted to Genbank), human (ZASP Accession number: AJ133766), and mouse Cypher2s (accession number AF114379). The sequence identities on the aa sequences are: mouse vs zebrafish 70.6%; human vs zebrafish 66.7%; and mouse vs human 92.4%. The exons are marked on top and correspond to the zebrafish gene. Shown in the middle is the alignment of exon 4 found exclusively in heart-specific splice variants. The sequence comparison of exons 4 and 6 is based on the genomic sequences of the different species. The ZM motif is marked on top, and mutations identified in human patients with myopathies are indicated by arrows below. Shown in the bottom is a similar alignment for exon 6 as shown above for exon 4. Arrows indicate mutations described for human patients with myopathies (red arrows refer to mutations described by Vatta et al., 2003 and black arrows indicate mutations described by Selcen and Engel, 2005). |
|
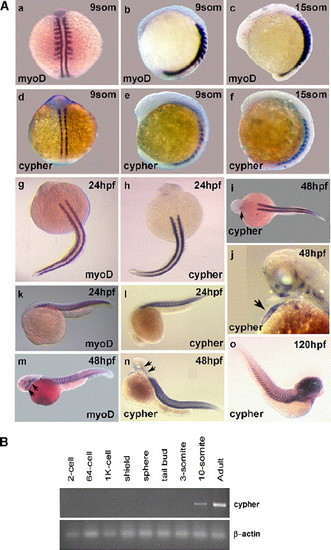
Temporal and spatial expression of cypher in developing zebrafish. (A) Shown are whole mount in situ hybridization results for the skeletal muscle-specific marker myoD and for cypher at different stages of zebrafish development. The earlier 9-somite (9som) stage (14 hpf) and 15-somite stage (about 17 hpf) are shown in a–f. Shown are dorsal and lateral views. Limited cypher expression was already observed at the 3, 5, and 7 somite stages (see Supplementary Fig. 1). Embryos at 24 h of development (g, h, k, and l), 48 h of development (i, j, m, and n), and 120 h of development (o) are shown below. Used was either a myoD or a cypher specific probe, as indicated. The arrows in i and j indicate staining in the heart and in m and n point to head and jaw musculature. Staining in the heart was observed for cypher at 36 hpf (see Supplementary Fig. 1) but not at 24 hpf (h and l). Myod staining was not present in the heart at all times. Expression for both cypher and myoD was detected in head and jaw musculature at 48 hpf (m, n and also j). Cypher expression in the brain was detected at 120 hpf (o). (B) Semi-quantitative RT-PCR using cypher2γ designed primers and RNA from different stages of zebrafish development standard RT-PCR protocols were used. The PCR was done with 28 cycles and β-actin was used to control quality and quantity of RNA used for the different embryonic stages. |
|
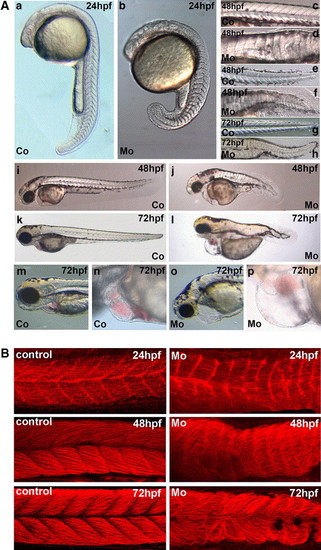
Effects of microinjection of cypher gene-specific morpholinos on zebrafish development. (A) Shown are stereoscope pictures and DIC microscopy pictures of embryos at different stages of development. The magnifications shown are in each case comparable between pictures of control and cypher-MO injected embryos but not for the different stages of embryos. The embryos were injected at the one-cell stage with either a control morpholino or the cypher specific morpholinos (2 ng embryo). a, c, e, g, i, k, m, and n show the control injected and b, d, f, h, j, l, o, and p correspond to the cypher-MO injected embryos. In a and b, embryos at 24 h of development are shown with (a) being the control and (b) the cypher-MO injected embryo. The picture in b of the cypher-MO injected embryo is focused on the somites and the notochord which is present in these embryos is not well visible. The notochord also in the later stages is malformed in cypher-MO injected embryos (d, f, h). Shown in c (control) and d (cypher-MO) are close ups of the region of the first 13 somites of embryos at 48 h of development. The somites of the tail region of 48 hpf embryos are shown in e (coer-MO injected embryos (d, f, h). Shown in c (control) and d (cypher-MO) are close ups of the region of the first 13 somites of embryntrol) and f (cypher-MO). g (control) and h (cypher-MO), the tail regions of 72 hpf embryos are shown. In i and j, embryos at 48 hpf and (k) and (l) at 72 hpf are shown. m and n show the head and heart of 72 hpf embryos of the control injected group whereas o and p show the cypher-MO injected group with enlarged pericardium. (B) Myotome boundaries in control and cypher-MO injected embryos. We used red phalloidin staining to detect F-actin which is present in all muscle cells. Shown on the left are control injected embryos and on the right cypher-MO injected embryos representing the severe phenotype. Time points used were 24 hpf, 48 hpf, and 72 hpf. The loss of organization and of myotome boundaries increases over time. (C) Somite staining in control and cypher-MO injected embryos. Embryos were injected with control (a-c) or cypher-MO (d-f) and fixed at the 15-somite stage. In situ hybridization for vmhc expression (-f) was performed and pictures from the dorsal view were taken. There are severe (f) and less severe deformed embryos (d, e) found. In g and h, in situ hybridization expression for myoD is shown, for a control injec PHENOTYPE:
|
|
Histology of zebrafish at 4 days of development. Shown are transversal sections of zebrafish at 4 days of development stained with toluidine blue. At the one-cell stage, zebrafish embryos were either injected with a control morpholino (control) or with the cypher-MO (MO). a and b show a comparison of more anterior somites of control and MO injected embryos. Large lacunae (La) are shown within the compartments of skeletal muscle in cypher-MO injected zebrafish (b and j). Transversal sections through more caudal regions are shown in c and d, these show the more posterior somites. g and h sections show the heart, with markedly enlarged and thinned pericardium (pc) and the thinned ventricular wall (vw) in the cypher-MO injected embryos (h). g and h are not absolutely comparable in scale in order to be able to show the enlarged pericardium in h. Shown in e and f are higher magnification pictures (40×) of the ventricles of either control or MO injected embryos, respectively. The ventricular wall (vw) is marked and is thinned in the cypher-MO injected embryos (f) compared to the control (e). Higher magnification (100× oil emersion) pictures of the anterior somites shown ir MO injected embryos, respectively. The ventricular wall (vw) is marked and is thinned in the cypher-MO injected embryos (f) compared to tr MO injected embryos, respectively. The ventricular wall (vw) is marked and is thinned in the cypher-MO injected embryos (f) compared to tn a and b are shown in i and j. Large lacunae (LA) in the m PHENOTYPE:
|
|
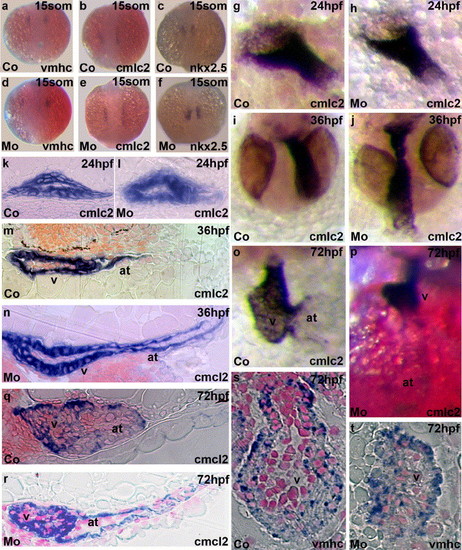
Effects of cypher gene-specific morpholinos on expression of heart development markers by in situ hybridization. Control injected embryos (a–c) and cypher-MO injected embryos (d–f) were fixed at the 15-somite stage and stained with probes for either ventricular myosin heavy chain (vmhc; a and d), cardiac myosin light chain 2 (cmlc2; b and e) or nkx2.5 (c and f). Hearts from control injected (g and k) or cypher-MO injected embryos (h and l) are shown at 24 hpf. Shown is the whole mount in situ staining for cmlc2. No significant differences in early heart development were observed in both the 15-somite embryos with the three different heart markers used and in the 24 hpf embryos stained with cmlc2. This is consistent with the late onset of cypher expression in the heart after 24 hpf. The earliest observation of a changed heart morphology was observed at 36 h of development, and cypher-MO injected embryos showed an elongated atrium (m and j) compared to the control (n and i). Shown in o–t are in situ hybridizations of 3 days post fertilization control injected embryos (o, q and s) and cypher-MO injected embryos (p, r, and t) for either cmlc2 (o–r) or vmhc (s and t) expression. Shown in q and r are longitudinal sections from 3 dpf embryos stained for cmlc2 and in s and t transversal sections of hearts from 3 dpf embryos stained for vmhc. Sections were counterstained with neutral red. The ventricle (v) and the atrium (at) are indicated in the figure. Similar results were found in all specimen investigated. EXPRESSION / LABELING:
PHENOTYPE:
|
|
Rescue of cypher-MO-induced phenotype. A rescue experiment as described in Table 2 was performed. Shown here are group pictures of embryos fixed at 3 dpf either injected at the 1-cell stage with cypher-MO (left; a; Mo, or injected with cypher-MO and 10 pg of cypher “rescue” mRNA (right; b; Mo + RNA). |
|
Effect of cyclopamine treatment on cypher gene expression. Shown in a, c, e, and g are control treated embryos. All other embryos were incubated with 50 μM cyclopamine (starting at shield stage). The minus and plus refer to cyclopamine treatment but only for the first row embryos, which show the dorsal view of somites at the indicated stages. The left panel of embryos (a–d, i, j, k, and o) was probed for myoD expression. myoD expression was examined as a control and to show the absence of myoD expressing adaxial slow muscle precursor cells in the somite stage embryos upon cyclopamine treatment (b and d). The right panel embryos (e–h, l, m, n, and p) were probed for cypher expression. Cypher expression was absent in 9-somite and 15-somite stage embryos treated with cyclopamine (f and h). Embryos at 24 h of development showed slightly reduced cypher expression in paraxial somites when treated with cyclopamine. This is more apparent in the dorsal view (p). Normal expression of cypher and myoD during embryogenesis is shown in Fig. 2A. The control treatment with ethanol had no effect on either cypher or myoD expression. Similar results were obtained in three independent experiments. EXPRESSION / LABELING:
|
|
Cypher expression shown by whole mount in situ hybridization. 3-somite stage (a), 5-somite stage (b), and 7-somite stage (c) embryos are shown in the top row. In (d), cypher expression is seen in the heart of the prim 25 stage (36 hpf) embryo. |
|
Cypher RNA was injected at the 1 cell stage at the indicated concentrations. An uninjected control, as well as embryos injected with 50ng were indistinguishable to the 100ng embryos. All did not show a phenotype. 500ng injected embryos showed severe defects and malformations already at early stage (14hpf). Toxicity is a possible explanation, or a combination of a specific effect and toxicity since the phenotypes induced were very severe and rather heterologous. EXPRESSION / LABELING:
|

Unillustrated author statements |
Reprinted from Developmental Biology, 299(2), van der Meer, D.L., Marques, I.J., Leito, J.T., Besser, J., Bakkers, J., Schoonheere, E., and Bagowski, C.P., Zebrafish cypher is important for somite formation and heart development, 356-372, Copyright (2006) with permission from Elsevier. Full text @ Dev. Biol.