- Title
-
Duplicate sfrp1 genes in zebrafish: sfrp1a is dynamically expressed in the developing central nervous system, gut and lateral line
- Authors
- Pezeron, G., Anselme, I., Laplante, M., Ellingsen, S., Becker, T.S., Rosa, F.M., Charnay, P., Schneider-Maunoury, S., Mourrain, P., and Ghislain, J.
- Source
- Full text @ Gene Expr. Patterns
|
Analysis of the enhancer trap line CLGY713 and identification of duplicate sfrp1 genes. Whole mount fluorescent imaging of live embryos (A–C) or YFP-immunostaining (D) of line CLGY713 was performed at the indicated stages. Organisation of the sfrp1a genomic locus (formerly sfrp1) and CLGY virus integration site (E). Amino acid sequence alignment of Sfrp1 proteins from zebrafish and mouse (F). The single letter code for amino acids is used. Amino acid numbering is indicated on the right. Identical and similar residues are indicated in black and grey, respectively. The conserved N-terminal cystein-rich domain (CRD) and C-terminal netrin (NTR) homology are shown. Phylogenetic tree of the full-length Sfrp1, Sfrp2 and Sfrp5 proteins (G). The degree of relatedness is indicated by the length of the vertical lines. Numbers indicate bootstrap support for the nodes. Ci, Ciona intestinalis; Dr, Danio rerio; hb, hindbrain; Hs, Homo sapiens; hpf, hours post-fertilisation; Mm, Mus musculus; ov, optic vesicle; ret, retina; ss, somite stage; teg, tegmentum; Tn, Tetraodon nigroviridis; Tr, Takifugu rubripes; vfb, ventral forebrain; 2°pllp, secondary posterior lateral line primordium. |
|
Analysis of sfrp1a expression during gastrulation and segmentation stages. Whole mount in situ hybridisation was performed with an antisense RNA probe for sfrp1a at the indicated stages (A–N). Double in situ hybridisations with antisense probes for pax2a (E, G and K), krx20 (F, H and J), pax6a (I) and wnt1 (L and M) are indicated and colour coded according to the substrate. In L and M, both wnt1 and sfrp1a probes were stained in blue. wnt1 marks the mhb, dorsal hindbrain (arrowhead in M) and is reinforced at inter-rhombomeric boundaries (black lines in L). Embryos are presented in whole mount (A–C, H, L and N), flat-mount (D–G and I–K) or a 40 μm transverse section of the hindbrain (M). Embryos are oriented with dorsal to the right (A and B) anterior up (C) or anterior to the left (D–L and N). amb, anterior midbrain; di, diencephalon; epi, epiboly; fb, forebrain; hb, hindbrain; lnp, lateral neural plate; mhb, mid-hindbrain boundary; ol, optic lobe; olf, olfactory placode; op, otic placode; ov, otic vesicle; pm, paraxial mesoderm; r, rhombomere; ret, retina; sh, shield; tb, tail-bud; teg, tegmentum. EXPRESSION / LABELING:
|
|
Analysis of sfrp1a expression in the developing gut. Whole mount in situ hybridisation was performed with an antisense RNA probe for sfrp1a (A, B, E and F) or fkd7 (C and D) at the indicated stages. Embryos are presented in whole mount with anterior to the left (A, C, E and F), or in 5 μm transverse sections (B and D) at the level indicated in A and C (white lines), respectively. A wild-type embryo is compared to a cas mutant embryo (E and F). The wild-type location of endodermal tissue is indicated with arrowheads. endo, endoderm; hyp, hypochord; pnd, pronephric duct. EXPRESSION / LABELING:
|
|
Analysis of sfrp1a expression in the developing lateral line. Whole mount in situ hybridisation was performed with an antisense RNA probe for sfrp1a at the indicated stages (A–H). A wild-type embryo is compared to a cls mutant embryo (G and H). Embryos are presented in whole mount with anterior to the left, in dorsal (A and C) or lateral view (B and D–H). all, anterior lateral line; fb, fin-bud; hb, hindbrain; inc, interneuromast cells; io, infraorbital lateral line; nm, neuromast; ov, otic vesicle; pll, posterior lateral line; pllp, posterior lateral line placode; pnm, proneuromast; ret, retina; so, supraorbital lateral line; 2°pllp, secondary posterior lateral line primordium. EXPRESSION / LABELING:
|
|
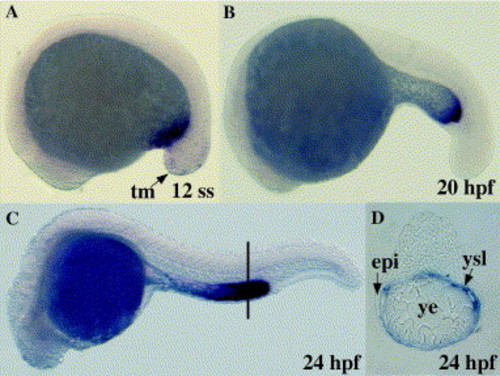
Analysis of sfrp1b expression during embryogenesis. Whole mount in situ hybridisation was performed with an antisense RNA probe for sfrp1b at the indicated stages (A–D). Embryos are presented in whole mount with anterior to the left (A–C) or in 5 μm transverse section (D) at the level indicated in C (black line). epi, epiderm; tm, tail mesenchyme; ysl, yolk syncytial layer; ye, yolk extension. EXPRESSION / LABELING:
|

Unillustrated author statements |
Reprinted from Gene expression patterns : GEP, 6(8), Pezeron, G., Anselme, I., Laplante, M., Ellingsen, S., Becker, T.S., Rosa, F.M., Charnay, P., Schneider-Maunoury, S., Mourrain, P., and Ghislain, J., Duplicate sfrp1 genes in zebrafish: sfrp1a is dynamically expressed in the developing central nervous system, gut and lateral line, 835-842, Copyright (2006) with permission from Elsevier. Full text @ Gene Expr. Patterns