- Title
-
Knockdown of zebrafish crim1 results in a bent tail phenotype with defects in somite and vascular development
- Authors
- Kinna, G., Kolle, G., Carter, A., Key, B., Lieschke, G.J., Perkins, A., and Little, M.H.
- Source
- Full text @ Mech. Dev.
|
Cloning and expression of zebrafish crim1. (A) Ideogram of the zebrafish Crim1 protein, showing domain homology (%) to human (H. sap), mouse (M. mus) and chicken (G. gal) Crim1 orthologs and C. elegans B0024.14 (C. eleg) proteins. S, signal peptide; I, insulin-like growth factor binding protein like domain; C1-6, Von Willebrand factor C type repeats; T, transmembrane domain; (B) crim1 positive signal was observed at non-saturation PCR conditions at all stages examined from blastula (256-1K cell) to 48 hpf. cDNA specific primers amplifying the ubiquitously expressed elongation factor-1 alpha (EF1α) was used as a loading control (Nordnes et al., 1994). (C-P) Expression of crim1 (C-P) at 16-cell (C), sphere (D), 50% epiboly (E), 75% epiboly (F), 90% epiboly (G), six-somite (H), 10-somite stage (I), 14-somite (J), 16-somite (K), 18-somite (L), 20-somite (M), 21 hpf (N), 24 hpf (O) and 30 hpf (P). All embryos are shown laterally with anterior to the top. In N, arrows denote anterior and posterior limits of crim1 expression in the notochord; in O and P, ICM; intermediate cell mass; O, otic vesicle; note the expanded ICM denoted by the arrow in B (severe panel). Inset in O (O′) is a dorsal view with anterior to the left of the embryo in O and represents the dorsal aorta (DA) and posterior cardinal vein (PCV). We note that the expression of crim1 in the brain ventricle (P) may be due to trapping of the riboprobe. Scale bar in C-N=250 μm; O and P=500 μm. |
|
Morpholino knockdown of zebrafish crim1. (A) Schematic showing the position of morpholino oligos designed across the start codon and (crim1A-MO) and to a region of the 5′ UTR (crim1B-MO) of zebrafish crim1 mRNA. To test the ability of crim1A-MO to knockdown expression of its target sequence, a Gfp reporter construct was produced. The start codon and first seven amino acids encode zebrafish crim1 sequence. (B) Brightfield and darkfield images showing that crim1A-MO but not a randomised control morpholino can block the production of GFP protein in zebrafish embryos injected with 500 pg of crim1-Gfp chaemeric mRNA. Scale bar=500 μm. (C) Graph showing the distribution of phenotypes observed by injection of 2.5 or 5 ng of control, crim1A-MO or crim1B-MO. Mild are embryos as represented in gray and severe in black. Standard error is deviation between separate injection experiments (n=4, 50–100 embryos/experiment). |
|
Analysis of crim1 morphants. (A) Graph showing percentages of phenotypes scored for crim1A-MO injected embryos at decreasing concentrations. (B) Representative images for the scored phenotypes of crim1A-MO injected embryos at 24–30 hpf. The pig-tail phenotype is described as having a twisted tail. Severely affected embryos have a grossly expanded intermediate cell mass (ICM) as shown by the black arrow. Unlike the ‘severe’ and ‘pig-tail’ phenotypes, morphants with the mild phenotype will often survive past 48 hpf. (C–D′). Zebrafish embryos injected with 5 ng of Control morpholino (C, D), or crim1A-MO (C′, D′) and photographed at 36 hpf (C, C′) and 48 hpf (D, D′). Morphants that survive to hatching stage develop the characteristic hook-tail (arrow in D′). Scale bar=100 μm. |
|
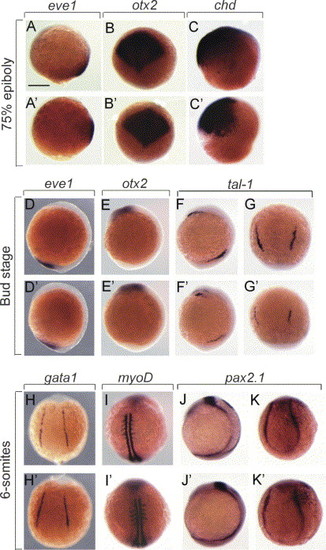
Analysis of dorsoventral markers on crim1 morphants. (A–K′). Wholemount RNA in situ hybridisation of 8–12 hpf embryos injected with 5 ng control-MO (A–K) and crim1A-MO (A′–K′) and stained with eve1 (A, A′, D, D′), otx2 (B, B′, E, E′), chordin (C, C′), tal1 (F, F′, G, G′), gata1 (H, H′), myoD (I, I′), and pax2.1 (J, J′, K, K′). Embryos are shown laterally with anterior to the top. Scale bar=300 μm. EXPRESSION / LABELING:
|
|
crim1 morphants have an expanded intermediate cell mass. (A–G′) Wholemount RNA in situ hybridisation of 24 hpf embryos injected with 5 ng crim1A-MO (B, E, F, H) and uninjected controls (A, C, E, G, I) and stained with tal1 (A, A′), lmo2 (B, B′), crim1 (C, C′), flk1 (D, D′), gdf6a/radar (E, E′), bmp4 (F, F′) and shh (G, G′). Arrows in A, A′, B′ and C′ denote the region of the intermediate cell mass (ICM). Note how the expression of these markers in the ICM is expanded in the crim1 morphants. It is also apparent that there are blood vessel defects as shown by the disorganised intersegmental vessels (Se) in D′ and major vessels in E′ (arrow). In pictures of the tail in G and G′, the inset (top right corner) shows the corresponding whole embryo. (H and H′). Whole mount immunohistochemistry of control-MO injected (G) and crim1A-MO injected (G′) embryos with En1 and visualized at 28 hpf. Embryos are shown laterally with anterior to the left. Se, intersegmental vessel; Scale bars=200 μm. EXPRESSION / LABELING:
|
|
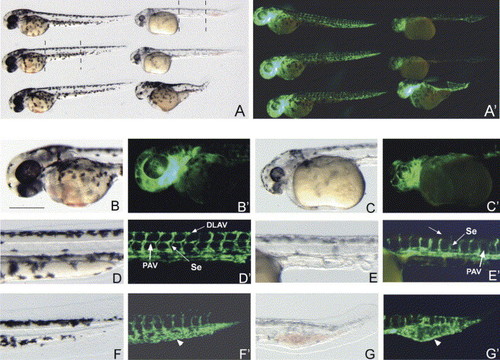
crim1 morphants display blood vessel defects. (A–G′). Brightfield and darkfield images of Control morpholino (B, B′, D, D′, F, F′) and crim1A-MO (C, C′, E, E′, G, G′) injected into transgenic fli:EGFP embryos and photographed post 48 hpf. In A and A′ panels, the embryos on the left are the Control morpholino injected embryos and on the right are the crim1A-MO injected embryos. Dotted lines in A represent section of embryos used in higher magnification. Note the reduction in GFP intensity in the crim1 morphants. The arrow in D′ shows the presence of the dorsal longitudinal anastomotic vessel (DLAV) and the parachordal vessels (PAV, white arrow) and their absence in E′. These panels (arrows-Se) also highlight the structural differences of the intersegmental vessels (Se) between the Control and crim1 morphants. Arrows in F′ and G′ point to the intermediate cell mass (ICM). Note how this region contains and is surrounded by GFP positive endothelial cells in the crim1 morphants. Scale bar=200 μm. |
Reprinted from Mechanisms of Development, 123(4), Kinna, G., Kolle, G., Carter, A., Key, B., Lieschke, G.J., Perkins, A., and Little, M.H., Knockdown of zebrafish crim1 results in a bent tail phenotype with defects in somite and vascular development, 277-287, Copyright (2006) with permission from Elsevier. Full text @ Mech. Dev.