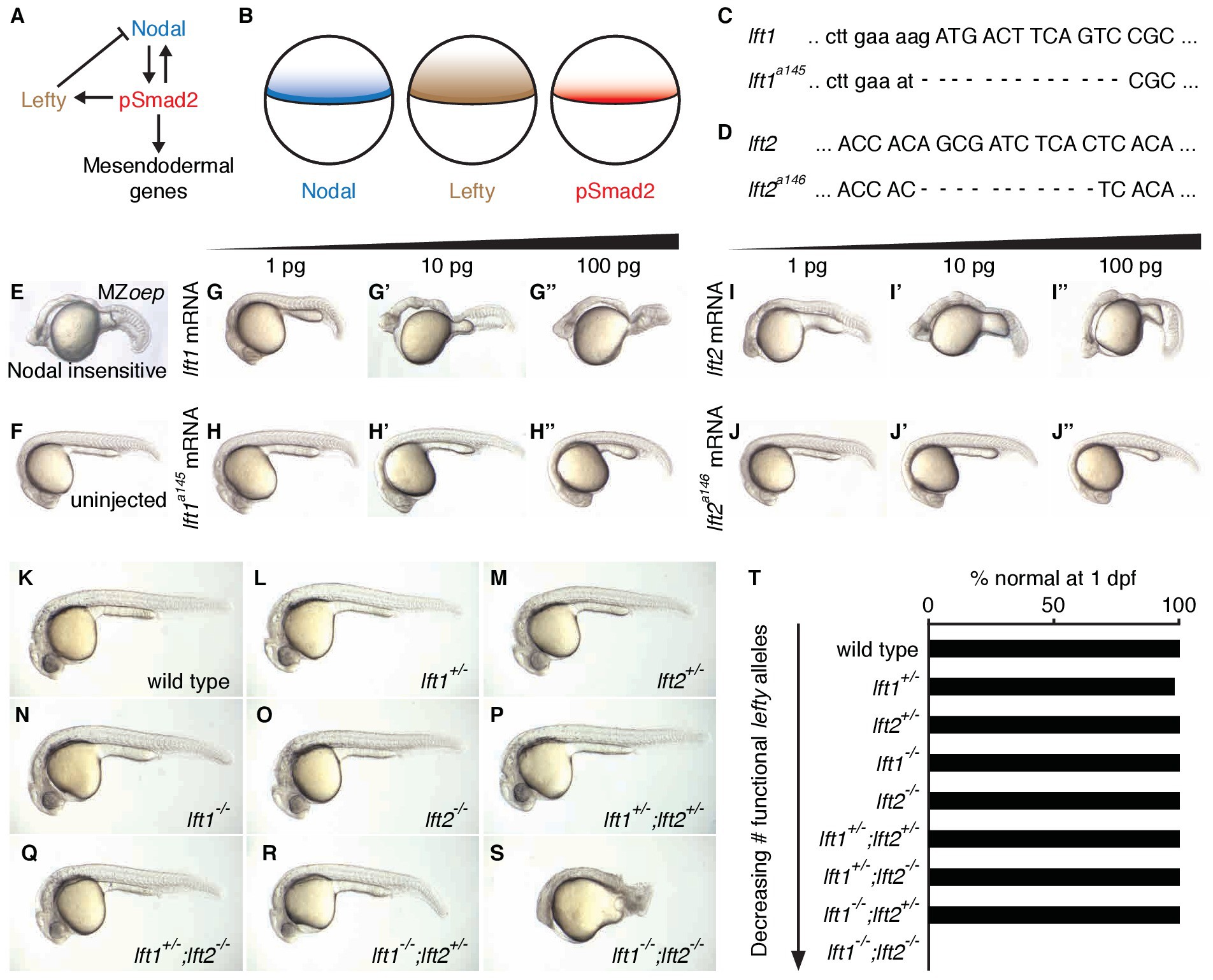
Fig. 1
Complete Lefty loss causes severe patterning defects.
(A) Nodal activates itself, mesendodermal genes, and the secreted feedback inhibitor Lefty by inducing phosphorylation and nuclear translocation of the signal transducer Smad2. (B) Nodal (blue) and Lefty (brown) are expressed in overlapping domains at the margin (black line) in zebrafish embryos, generating a signaling gradient of phosphorylated Smad2 (red). (C) A 13-base-pair deletion at the 5’ end of lft1a145 removes the translational start site (TSS) and part of the signal sequence. An alternative TSS 36 bp downstream of the deletion could produce an in-frame protein product, but the first 16 amino acids, and therefore most of the predicted 20-aa signal sequence, would be missing. (D) An 11-base-pair deletion at the 5’ end of lft2a146 removes part of the predicted 19-aa signal sequence, causing a frame shift after 36 bp resulting in a stop codon 18 bp later. (E–J’’) Testing the activity of lft mutant mRNA. All images were obtained at 24 hr post-fertilization (hpf). Wild type embryos at the one-cell stage were injected with 1, 10, and 100 pg wild type lft1 (G–G’’) or lft1a145 (H–H’’) mRNA as indicated. Embryos expressing lft1 mRNA exhibit Nodal loss-of-function phenotypes, similar to maternal-zygotic mutants for the zebrafish EGF-CFC co-receptor one-eyed pinhead (oep) (E), which are insensitive to Nodal signaling (Gritsman et al., 1999). Embryos expressing mutant mRNA do not exhibit Nodal loss-of-function phenotypes. Similar results were obtained for wild type lft2 (I–I’’) and lft2a146 (J–J’’) mRNA. (F) Uninjected wild type embryo. (K–S) lft mutant phenotypes. All images were obtained at 24–29 hpf. A single functional lft allele is sufficient for grossly normal patterning (Q,R). (S) lft1-/-;lft2-/- double homozygous mutants have severe patterning defects and lack eyes, heart, and full length tails, and often exhibit excess tissue along the posterior trunk. (T) Percentage of embryos with normal gross morphology at 1 day post-fertilization (dpf). Number of normal/total embryos: wild type = 50/50, lft1+/- = 49/50, lft2+/- = 46/46, lft1-/- = 50/50, lft2-/- = 46/46, lft1+/-;lft2+/- = 50/50, lft1+/-;lft2-/- = 43/43, lft1-/-;lft2+/- = 24/24 lft1-/-;lft2-/- = 0/55.