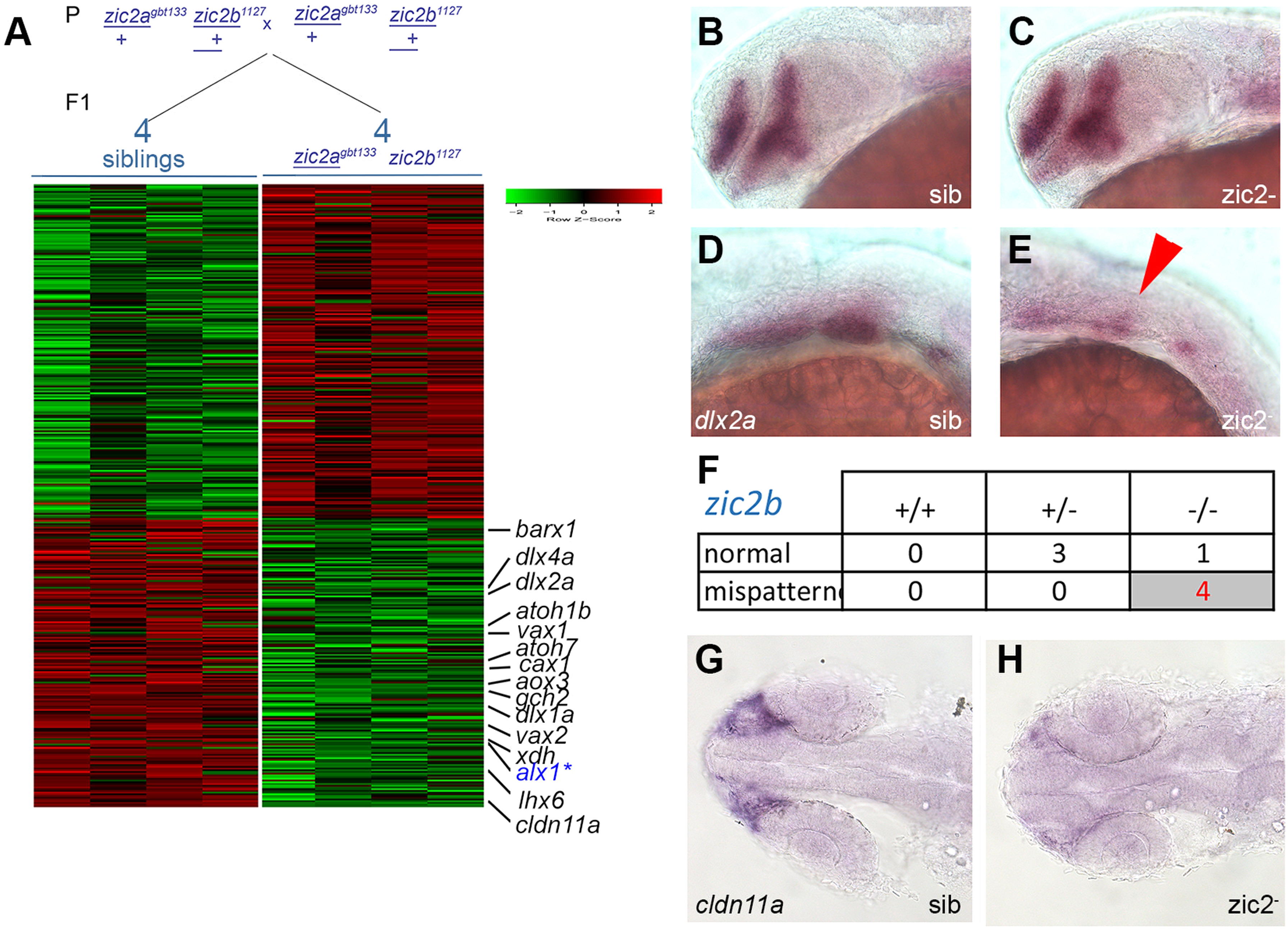
Fig. 5
RNA sequencing transcriptome analysis identifies a set of Zic2-dependent targets. A: Embryos derived from zic2agbt133/+; zic2b1127/+ parents were sorted by presence or absence of coloboma into zic2 mutants and sibling groups, respectively. RNA extracted from individual embryos (4 wildtype and 5 with coloboma) was used to prepare cDNA libraries for illumina high-throughput sequencing. Genes with assigned value of False-Discovery Rate below 0.05 were preliminarily selected. The heat-map color represents relative expression levels of differentially expressed genes; 4 out of 5 coloboma-representing libraries are shown to maintain visual balance with the 4 normal sibling samples. B, C: Representative sibling and zic2 mutant embryos derived from zic2agbt133/+;zic2buw1116/+ parents show normal dlx2a expression by WISH in the telencephalon and diencephalon. D: normal dlx2a expression in branchial arch primordia. E: depleted branchial arch dlx2a expression (arrowhead) was observed in 7 out of 127 (6%, 3 expts.) of embryos from this cross. F: Only zic2b- homozygotes exhibited dlx2a reduction in branchial arch primordia. G: normal cldn11a expression adjacent to the optic stalk of embryo with normal retinal morphology. H: depleted cldn11a expression in zic2 mutant with coloboma. Embryos in B – E are shown in lateral views, anterior to the left. Embryos in G and H are shown in dorsal views, anterior to the left.
Reprinted from Developmental Biology, 429(1), Sedykh, I., Yoon, B., Roberson, L., Moskvin, O., Dewey, C.N., Grinblat, Y., Zebrafish zic2 controls formation of periocular neural crest and choroid fissure morphogenesis, 92-104, Copyright (2017) with permission from Elsevier. Full text @ Dev. Biol.