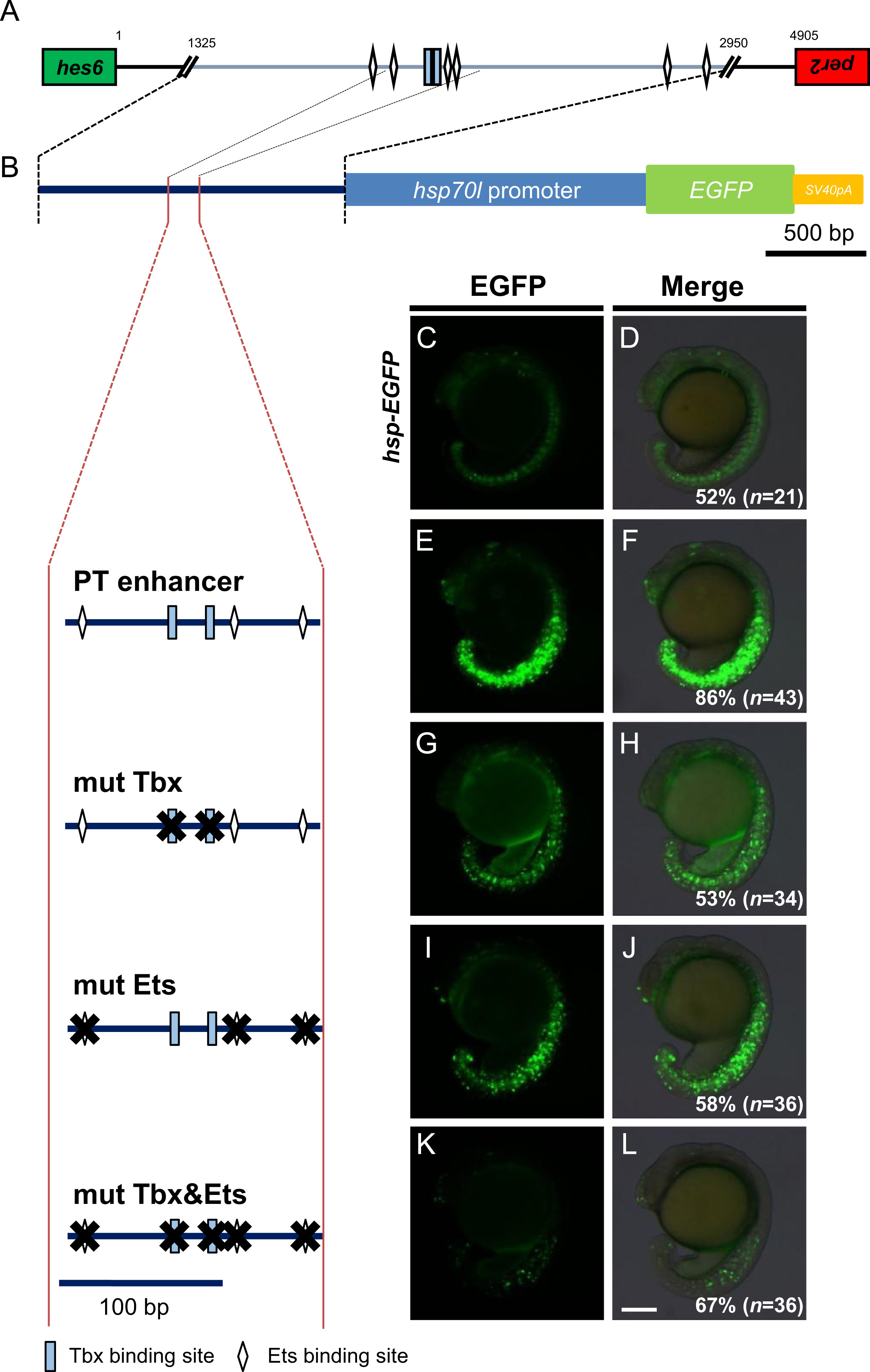
Fig. 3
Binding sites of T-box and Ets transcription factors are required for transcriptional regulatory activities of the PT enhancer. (A) Cluster of putative T-box and Ets transcription factor binding sites in the PT enhancer identified using the Patch 1.0 program. Rectangles represent T-box binding motifs. Diamonds indicate putative Ets binding sites. The green box represents the last exon of hes6, whereas the red box is the last exon of per2. The orientation of transcription in hes6 and per2 is the tail-to-tail opposite in zebrafish. The numbers shown above the bar indicates the base pair distance from the first intergenic sequence between hes6 and per2. (B) The mutations introduced into the T-box and/or Ets binding sites in the PT enhancer ligated to the hsp70l->EGFP reporter (shown at top) are shown at left. Several mutated reporter genes were injected into fertilized eggs at a concentration of 10 ng/µl and the EGFP signals in the PSM and tailbud were examined at the 15–18-somite stage, shown at right. The percentage of embryos showing the expression patterns of EGFP shown in the panels relative to those of injected normal embryos (n) is shown at the bottom-right of each panel. All images were taken at the same magnification. Scale bar, 200 µm.
Reprinted from Developmental Biology, 409(2), Kawamura, A., Ovara, H., Ooka, Y., Kinoshita, H., Hoshikawa, M., Nakajo, K., Yokota, D., Fujino, Y., Higashijima, S.I., Takada, S., Yamasu, K., Posterior-anterior gradient of zebrafish hes6 expression in the presomitic mesoderm is established by the combinatorial functions of the downstream enhancer and 3'UTR, 543-54, Copyright (2016) with permission from Elsevier. Full text @ Dev. Biol.