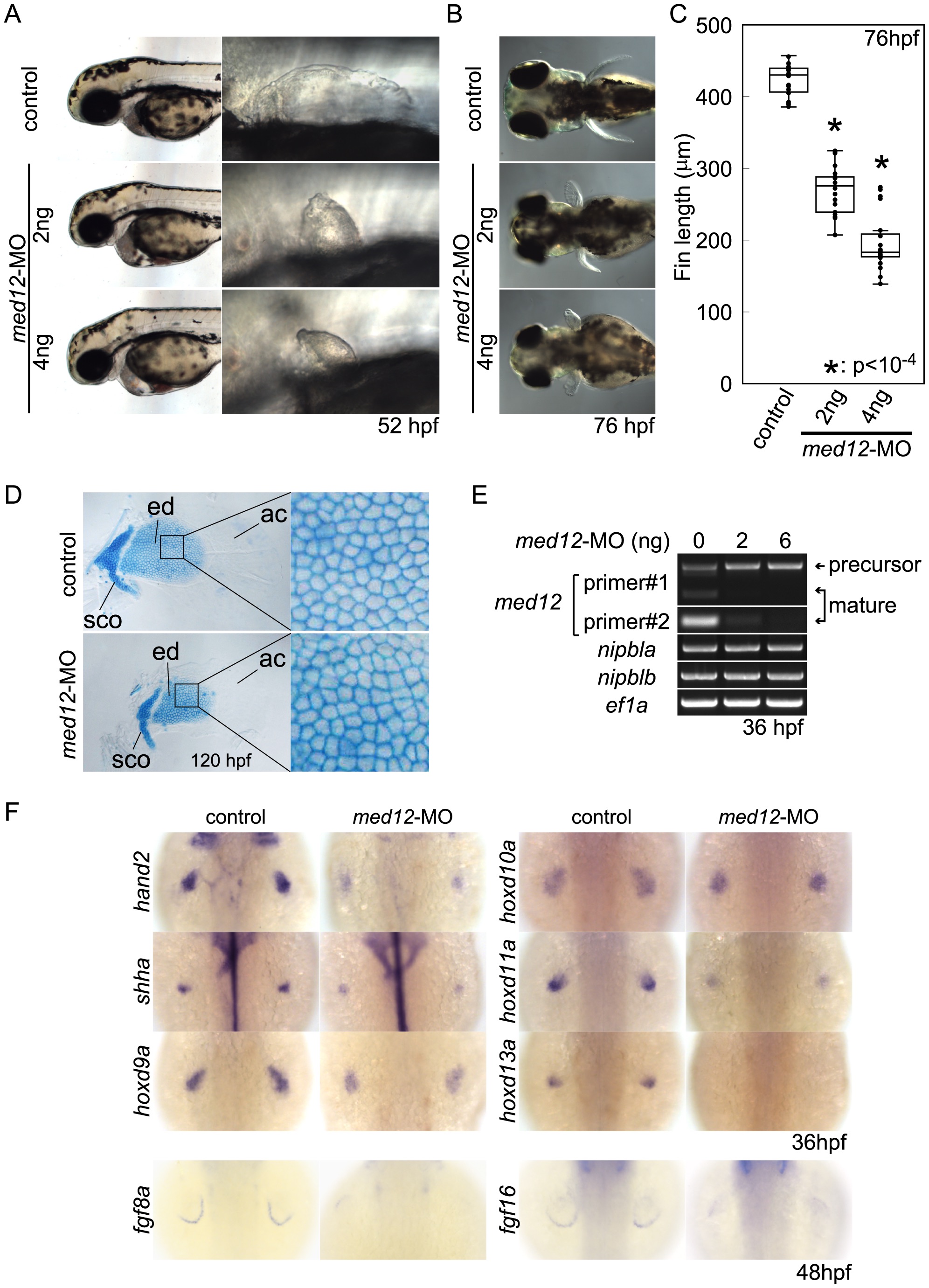
Fig. 7
Fig. 7 Med12 depletion disrupts pectoral fin morphology and gene expression similar to Nipbl depletion.
(A, B) Morphology of live embryos at 52 hpf (A, lateral views) and 76 hpf (B, dorsal views). (A) Anterior halves of control and med12-MO-injected embryos (left column) and higher magnification pictures of their pectoral fin buds (right column). (B) Dorsal views of embryos at 76 hpf. (C) Whisker plots of fin length. Fin lengths (medians) are 430.0 μm, n = 18 (control), 275.6 μm, n = 20 (med12-MO, 2 ng), and 183.8 μm, n = 20 (med12-MO, 4 ng). *: p<10-4. (D) Alcian blue staining of pectoral fin cartilage of control (upper) and Med12-deficient (med12-MO, 4 ng; lower) embryos at 120 hpf. Dorsal view, anterior to the top. Right column, higher magnification pictures of boxed areas of endoskeletal discs. ac, actinotrichs; ed, endoskeletal disc; sco, scapulocoracoid. (E) Controls for med12-MO efficiency. RT-PCR, 36 hpf. Both pairs of med12 primers (Primer #1 and #2) show that splicing of med12 mRNA is significantly suppressed by med12-MO, with a slightly higher efficiency at 6 ng. Primer pair #1 detects both precursor and mature mRNA, whereas primer pair #2 only detects mature mRNA (see Materials and Methods). nipbla and nipblb expression was not affected by Med12 depletion. ef1a was used as a control. (F) Expression of genes involved in the 5′-hox/hand2/shha gene cassette and AER fgf genes in pectoral fin buds examined by ISH at 36 hpf. Dorsal views, anterior to the top. Similar to Nipbl-deficient embryos, shha, hand2 and 5′-hoxd genes in mesenchyme as well as fgf16 and fgf8a in the AER are reduced in Med12-deficient embryos (4 ng/embryo med12-MO).