Fig. 5
- ID
- ZDB-FIG-161229-17
- Publication
- Venero Galanternik M. et al., 2016 - Glypican4 Modulates Lateral Line Collective Cell Migration Non Cell-Autonomously
- Other Figures
- All Figure Page
- Back to All Figure Page
|
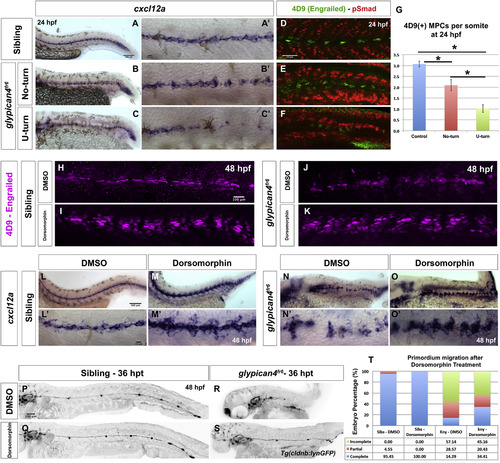
Bmp inhibition rescues Engrailed(+) muscle cells and cxcl12a expression in glypican4fr6. (A-C′) In situ hybridization shows expression of the Chemokine ligand cxcl12a in siblings (A-A′), glypican4fr6 No-turning mutants (B-B′) and glypican4fr6 U-turning mutants (C-C′) along the embryo trunk. (A′-C′) Higher magnification images of (A-C). (d-F) Engrailed (4D9) immunostaining reveals that, compared to siblings (D), glypican4fr6 No-turning mutants (E) and glypican4fr6 U-turning mutants (F) have a considerably diminished number of MPCs and MFFs. (G) Quantification of 4D9-positive muscle pioneer cells per somite at 24 hpf (p-values (t-test) for Control vs. No-turn=0.0036, Control vs. U-turn=5.09721×10−12, No-turn vs. U-turn=0.00065). Error bars represent standard error. (H-K) In control (H-I) and glypican4fr6 mutant (J-K) embryos inhibition of Bmp signaling with 10 μM Dorsomorphin induces the formation of MPCs and MFFs along the myoseptum. (L-O′) Bmp inhibition results in the upregulation of cxcl12a expression along the myoseptum in siblings (L-M′) and restores cxcl12a expression in glypican4fr6 mutants (N-O′) along the embryo trunk. (L′-O′) Higher magnification images of (L-O). (P-T) Inhibition of Bmp signaling does not affect primordium migration (P-Q) but significantly rescues the migration of glypican4fr6 mutants (R-S). (T) Quantification of primordia that have completed migration after Bmp inhibition, Fisher Exact Probability Test, Sibling-DMSO vs. Siblings-Dorsomorphin =0.31; glypican4fr6-DMSO vs. glypican4fr6-Dorsomorphin=0.04. Hours post treatment (hpt). |
| Genes: | |
|---|---|
| Antibody: | |
| Fish: | |
| Condition: | |
| Anatomical Terms: | |
| Stage Range: | Prim-5 to Long-pec |
| Fish: | |
|---|---|
| Condition: | |
| Observed In: | |
| Stage Range: | Prim-5 to Long-pec |
Reprinted from Developmental Biology, 419(2), Venero Galanternik M., Lush, M.E., Piotrowski, T., Glypican4 Modulates Lateral Line Collective Cell Migration Non Cell-Autonomously, 321-335, Copyright (2016) with permission from Elsevier. Full text @ Dev. Biol.