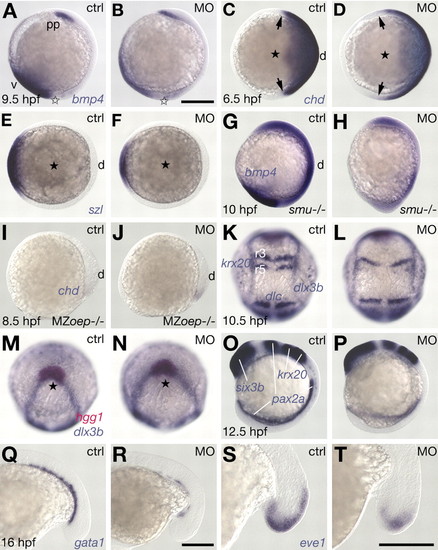
MO-induced interference with Prdm1 activity affects DV patterning. (A,B,G-J,O,P) Lateral view, dorsal to the right. (C-F) Animal view, dorsal to the right. (K,L) Dorsal view, anterior to the top. (M,N) Animal view, dorsal to the bottom. (K,M;L,N) different views of same embryos. (Q-T) Lateral views, anterior to the left; tailbud region is shown. (A,C,E,G,I,K,M,O,Q,S) Untreated controls (ctrl). (B,D,F,H,J,L,N,P) Embryos microinjected with 2 ng of MOprdm1 (MO). (R,T) Embryos microinjected with 1 ng of MOprdm1. (A,B) bmp4 expression. (C,D) chd expression, domain boundaries highlighted (arrows). (E,F) szl expression. (G,H) Reduced ventral expression of bmp4 in morphant smu embryos; lack of ptc1 expression identifies smu mutants (not shown). (I,J) Increased dorsal chd expression in MZoep mutant embryos. (K,L) Mediolateral expansion of forming neural and somitic tissues as revealed by expression of krox20, dlx3b and dlc. (M,N) Formation of the prechordal plate was not altered in morphants. (O,P) AP neural patterning, revealed by six3b, pax2a and krox20 expression. (Q,R) Ventroposterior expression of gata1 in blood precursors. (S,T) Expression of eve1 in ventroposterior cells in the tail tip at midsegmentation. Scale bar: 200 µm. d, dorsal; r3/rS, rhombomeres 3 and 5; v, ventral. Animal pole highlighted by a black star. The vegetal pole is highlighted by a white star.
|

